Fasting (Python Package)¶
Make use of your fasting logs.¶
This tutorial provides a brief overview of how to use the fasting library. We'll use a sample data export from Zero in the tutorial.
This package is part of the Digital Biomarker Discovery Pipeline (DBDP). You can read more about the DBDP here. If you track your fasts, there's a chance you also track biomarkers with wearables and other hardware in the modern health stack. The goal of this package is to enable you to utilize your data for your own purpose, like cross analyzing fasting performance with other biomarkers. Check out an example here.
Fasting¶
Fasting is a technique used to improve metabolic health, reduce inflammation, rebalance gut health, lose weight, increase longevity, improve mental clarity and focus.
There's various forms of fasting, with Intermittent Fasting (IF) likely being the most common. You can read about IF and other types of fasting on the Zero blog.
Getting Started¶
Let's start by importing the packages we'll need for this tutorial.
from fasting import quantify
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
Data¶
Zero provides a data export feature in their app. This dataset is a sample users can expect to see from their own export.
Exporting Data from Zero¶
The data export feature in Zero can be found in the My Data section in the me page settings. To export your own data:
- Me (lower right) -> Settings (upper right) -> My Data -> Export Data
Loading the Data¶
First we'll load the raw data export into a Pandas DataFrame to better understand why we need to use the fasting library.
export = "https://raw.githubusercontent.com/jbpauly/fasting/main/docs/tutorials/zero_export.csv"
zero_export = pd.read_csv(export)
zero_export[-5:]
| Date | Start | End | Hours | Night Eating | |
|---|---|---|---|---|---|
| 24 | 1/18/21 | 19:14 | 11:44 | 16 | 1 |
| 25 | 1/17/21 | 19:15 | 12:00 | 16 | 1 |
| 26 | 1/16/21 | 19:45 | 10:14 | 14 | 2 |
| 27 | 1/15/21 | 21:00 | 10:00 | 13 | 3 |
| 28 | 1/14/21 | 19:00 | 11:22 | 16 | 1 |
Here we can see the raw data export provides sufficient data around each fast a user participated in through the Zero app.
- Date: Start date of the fast
- Start: Start time of the fast
- End: End time of the fast
- Hours: Duration of the fast rounded to the nearest hour
- Night Eating: Duration of time between sunset and fast start in hours
Alone, hours and night eating are usable metrics, but there's much more to gather out of our dataset. To do so, we'll simplify the logs to a start datetime and end datetime for each fast by combining date, start, end, hours.
Discrete Logs: Start and End Datetime¶
To create the discrete logs, lets read in the data using the zero_fasts function in the quantify module.
fasts_discrete = quantify.zero_fasts(export)
fasts_discrete[:5]
| start_dt | end_dt | |
|---|---|---|
| 0 | 2021-01-14 19:00:00 | 2021-01-15 11:22:00 |
| 1 | 2021-01-15 21:00:00 | 2021-01-16 10:00:00 |
| 2 | 2021-01-16 19:45:00 | 2021-01-17 10:14:00 |
| 3 | 2021-01-17 19:15:00 | 2021-01-18 12:00:00 |
| 4 | 2021-01-18 19:14:00 | 2021-01-19 11:44:00 |
We now have a start and end datetime (start_dt, end_dt) to work with for each fast. Additionally, the fasts are reindexed from oldest to newest, and any incomplete or ongoing fasts have been dropped.
Why start and end datetime?¶
Because fasting is primarily a time based protocol, it's important to get our data into a datetime format. From here we can take it one step further from discrete logs with start and end datetimes to a continuous time series log. Time series data can be tricky to handle. To overcome many challenges working with time based data, it's useful to work with it as a time series.
Continuous Log: Time Series of Fasting Status (not fasting/fasting)¶
Lets now convert the the discrete logs to a single continuous log using the continuous_fasts function in the quantify module. This function reads in the discrete fasts as a Pandas DataFrame and returns a binary Pandas Series:
- 0: Not fasting
- 1: Fasting
The series duration spans from the start_dt of the first fast in the discrete fasts log to the end_dt of the last fast in the discrete fasts log and has a sample rate of one minute.
fasts_continuous = quantify.continuous_fasts(fasts_discrete)
fasts_continuous[:5]
2021-01-14 19:00:00 1 2021-01-14 19:01:00 1 2021-01-14 19:02:00 1 2021-01-14 19:03:00 1 2021-01-14 19:04:00 1 Freq: T, dtype: int64
It's now less useful to tabularize the data. Instead, we can visualize it.
fig, ax = plt.subplots(figsize=(12,6))
ax.plot(fasts_continuous)
fig.autofmt_xdate()
plt.title('Binary Fasting Log: 0 (Not Fasting), 1 (Fasting)')
plt.xlabel('Date')
plt.ylabel('Fasting Status')
ax.set_yticks([0,1])
plt.show()
Why a continuous time series?¶
Pandas and other Python libraries support a vast array of time series functionality. To utilize these libraries, we often need our data as a series to perform aggregations, windowing functions, etc.
For data storage, it may be best to write discrete logs to a database instead of a continuous log. Two data points (start and end datetime) is much less than the thousands of points in a single fast continuous log.
Fasting Metrics¶
With the continuous log, we can now calculate various metrics of interest around our fasts.
Continuous Metrics¶
There may be some instances where it's useful to stay in the continuous space, like calculating the cumulative sum (consecutive) minutes of each fast.
consecutive_minutes = quantify.consecutive_minutes(fasts_continuous)
consecutive_minutes[:5]
2021-01-14 19:00:00 1 2021-01-14 19:01:00 2 2021-01-14 19:02:00 3 2021-01-14 19:03:00 4 2021-01-14 19:04:00 5 Freq: T, dtype: int64
fig, ax = plt.subplots(figsize=(12,6))
ax.plot(consecutive_minutes)
fig.autofmt_xdate()
plt.title('Running Consecutive Minutes of Fasting')
plt.xlabel('Date')
plt.ylabel('Minutes')
plt.show()
With the consecutive minutes, you could label each time step with the associated fasting zone*:
- Anabolic (0-4 hours)
- Catabolic (4-16 hours)
- Fat Burning (16-24 hours)
- Ketosis (24-72 hours)
- Deep Ketosis (72+ hours)
*Note: this can be provided as a function in future versions of the package.
 Unique physiologic effects occur within each fasting zone. You can learn more about the physiology of fasting from Zero here.
Unique physiologic effects occur within each fasting zone. You can learn more about the physiology of fasting from Zero here.
Discrete Metrics¶
Often times, biometrics are rolled up to discrete metrics at a specified frequency (i.e. daily). Since fasts can span across multiple days and multiple fasts can occur in a single day, we have to be careful how we roll up our discrete metrics. The image below helps explain calculations like cumulative hours and maximum consecutive hours fasted.
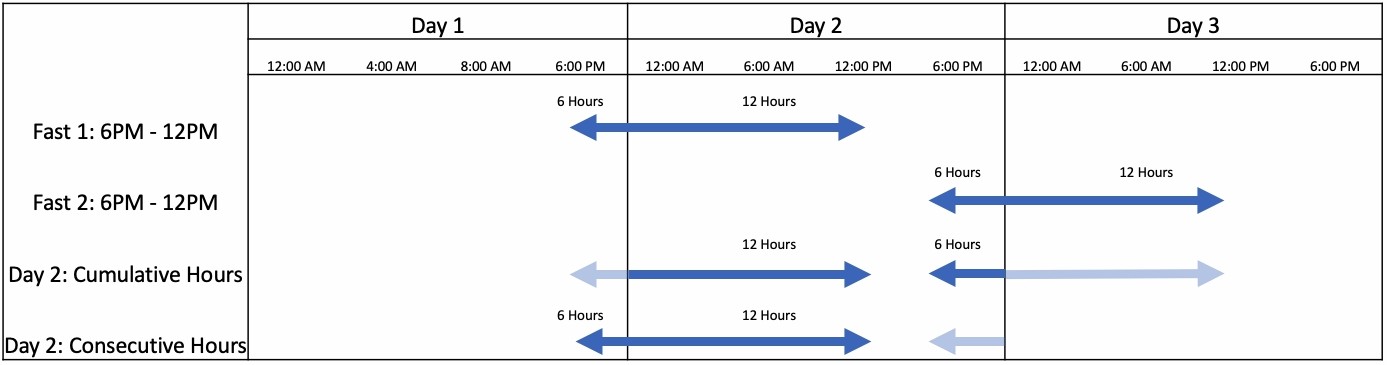
This diagram also reiterates the importance of working with time data as a time series. Even though we are calculating discrete metrics, it's easier to go from a higher data space (continuous log) down to a lower data space (daily metrics).
Cumulative Hours¶
Simply the cumulative hours fasting each day. Since the continuous log is a binary series of 0's and 1's, we can sum the series at a daily frequency.
cumulative_hours = quantify.daily_cumulative_hours(fasts_continuous)
cumulative_hours[:5]
2021-01-14 5.000000 2021-01-15 14.383333 2021-01-16 14.266667 2021-01-17 15.000000 2021-01-18 16.783333 Freq: D, dtype: float64
fig, ax = plt.subplots(figsize=(12,6))
ax.bar(cumulative_hours.index, cumulative_hours.values)
fig.autofmt_xdate()
plt.title('Cumulative Hours of Fasting per Day')
plt.xlabel('Date')
plt.ylabel('Hours')
plt.show()
Maximum Consecutive Hours¶
You may participate in multiple fasts each day. Maybe you complete one fast at 12PM with lunch and start your next fast at 7PM after dinner. The maximum consecutive hours is the maximum consecutive hours fasted within each day. It can included hours rolled over from the previous day(s).
max_consecutive_hours = quantify.daily_max_consecutive_hours(fasts_continuous)
max_consecutive_hours[:5]
2021-01-14 5.000000 2021-01-15 16.383333 2021-01-16 13.016667 2021-01-17 14.500000 2021-01-18 16.766667 Freq: D, dtype: float64
fig, ax = plt.subplots(figsize=(12,6))
ax.bar(max_consecutive_hours.index, max_consecutive_hours.values)
fig.autofmt_xdate()
plt.title('Maximum Consecutive Hours of Fasting per Day')
plt.xlabel('Date')
plt.ylabel('Hours')
plt.show()